(last modified March 5, 2006)
The DMD gene produces a range of different transcripts encoding various dystrophin isoforms, i.e. proteins of varying lengths containing different segments of the basic dystrophin sequence. The isoforms are encoded by a range of different mRNA's which are generated by three processes;
Depending on the technique used, it can be very difficult to discriminate between all dystrophin isoforms. Antibodies will detect several isoforms in immuno-histochemistry and primer pairs will amplify several isoforms in RT-PCR. Every time a new isoform is identified, one should check if previous experiments were conclusive in discriminating between an isoform studied and the newly detected isoform. Throughout these pages, we have used a specific nomenclature to describe the individual dystrophin isoforms.
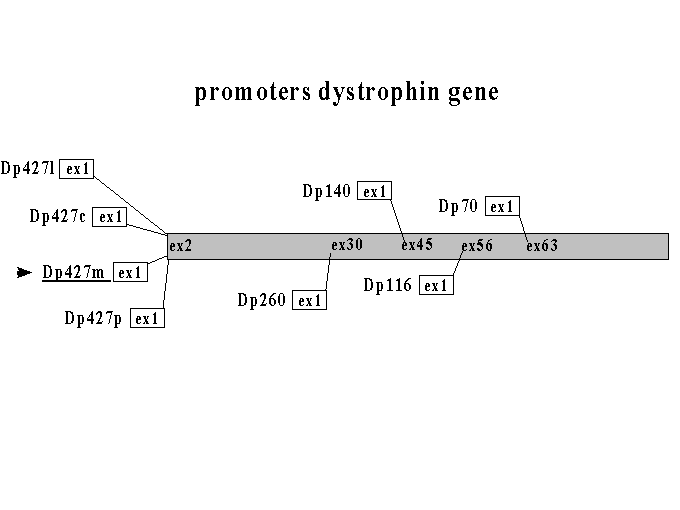
For clarity, we describe the different DMD/dystrophin proteins (Dp) throughout these pages in a specific manner, which is based on their length in kiloDaltons (Dp427, Dp260, etc.), the tissue of expression (Dp427m from muscle, Dp427p from Purkinje cells, etc.) and the differential splicing pattern generating them (Dp71a, Dp71b, etc.).
Analysis of dystrophin sequences in lower eukaryotes shows that the most ancient form of dystrophin is the one without the human exon 78 sequence (Wang et al. [1998]). As a consequence, the last 13 amino acids (RNTPGKPMREDTM) of the full length DMD-protein are replaced by 31 new amino acids (HNVGSLFHMADDLGRAMESLVSVMTDEEGAE). These latter amino aicds are conserved down to insects.
Most of the different transcripts (isoforms) of the dystrophin gene are evolutionary conserved (mammals). Analysis of dystrophin transcripts in the fruit fly (Drosophila) shows that the presence of several independent promoters driving transcription of the DMD gene is evolutionairy conserved, dating back to before the divergence between protostomes and deuterostomes.
| Name | synoniem | synoniem | protein length | amino acids | mRNA | promoter located in | expression | Reference |
|---|---|---|---|---|---|---|---|---|
| Dp427l | lymphocyte dystrophin | L-dystrophin | 427 kDa | 3,562 | 13,764 bp | 5' Dp427c | lymphoblastoid | Nishio, Muntoni, Wheway & Roberts |
| Dp427c | cortical dystrophin | brain or C-dystrophin | 427 kDa | 3,677 | 14,069 bp | 5' Dp427m | brain | Nudel |
| Dp427m | muscle dystrophin | M-dystrophin | 427 kDa | 3,685 | 13,993 bp | 5' of gene | muscle | Monaco, Koenig |
| Dp427p | Purkinje dystrophin | P-dystrophin | 427 kDa | 3,681 | 14 kb | 3' Dp427m | Purkinje cells | Gorecki, Holder |
| fetal dystrophin | dystrophin-delC | Feener | ||||||
| Dp260 | retinal dystrophin | R-dystrophin | 260 kDa | intron 29 | retina | D'Souza | ||
| Dp260-1 | R-1 | 2,344 | 9,773 bp | retina | White,R.A. U27203 | |||
| Dp260-2 | R-2 | 2,341 | 9,916 bp | retina | White,R.A. U27203 | |||
| Dp140 | 140 kDa | 1,225 | 7,410 bp | intron 44 | central nervus system and kidney | Lidov, Lidov | ||
| Dp140b | 1,243 | 7,378 bp | kidney | Lidov | ||||
| Dp140ab | 1,230 | 7,339 bp | cerebellum and kidney | Lidov | ||||
| Dp140c | 1,115 | 7,050 bp | cerebellum | Lidov | ||||
| Dp140bc | 1,133 | 7,048 bp | cerebellum and kidney | Lidov | ||||
| Dp116 | apo-dystrophin 2 | S-dystrophin | 116 kDa | 956 | 5,623 bp | intron 55 | Schwann cells | Byers |
| Dp71 | apo-dystrophin 1 | liver or G-dystrophin | 71 kDa (70.4) | 617 | 4,623 bp | intron 62 | ubiquitous | Lederfein, Austin |
| Dp71b | 72.2 kDa | 635 | 4,591 bp | ubiquitous | Austin | |||
| Dp71a | 68.9 kDa | 604 | 4,584 bp | ubiquitous | Austin | |||
| Dp71ab | 70.8 kDa | 622 | 4,552 bp | ubiquitous | Austin | |||
| Dp40 | apo-dystrophin 3 | 40 kDa | 340 | 2.2 kb | intron 62 | ubiquitous | Tinsley |
Legend:
Name: abbreviation used throughout these pages. a-types miss exon 71, b-types
miss exon 78, ab-types exons 71 and 78, c-types exons 71-74, etc. Synoniem:
alternative names used in literature. Protein length: length of the dystrophin
isoform in kiloDalton. Amino acids: protein length in amino acids. mRNA :
lenght of the mRNA in kilo basepairs. Expression: tissues in which the isoform has
been reported to be expressed (major expression in bold). Reference: publications
containing details regarding the dystrophin isoform.

The main dystrophin found in muscle is designated Dp427m. Its exon 1 encodes a unique N-terminal MLWWEEVEDCY amino acid sequence.
Dp427m is expressed in skeletal muscle, heart ...
This isoform, described first by Nishio et al. (1994), was found in lymphoblastoid cells. Transcripts originate at a unique promoter/exon 1 (Dp427l), located centromeric of DXS84 (754), and spliced to exon 3 of the normal Dp427m transcript (see below). As a consequence amino acids 1-31 are replaced by a single Methionine. The promoter/exon 1 region of this transcript shows a 80% homology to MER17A1 repetitive DNA. The Dp427l isoform is also known as L-dystrophin.
The presence of this isoform was confirmed by Muntoni et al. (1995), although these authors report splicing to dystrophin exon 2 only. This observation questions the reported translational start of Dp427l since splicing to exon 2 would not create an open reading frame before the Methionine at amino acid 124 of Dp427m (in exon 6).
Wheway & Roberts (2003) recently re-evaluated the Dp427l promoter in the light of available human and mouse genome sequences and attempted to compare its activity in lymphocytes with that of the Dp427m promoter. Genome data show that the Dp427l promoter lies immediately adjacent to the CGD (chronic granulomatous disease) gene (CYBB), i.e ~4.5 Mb upstream of DMD exon 2. This would make the Dp427l transcript 7 Mb in size, giving an estimated time for transcription of over 2 days. Transcription from the Dp427l promoter in peripheral blood lymphocytes was negligible compared to that of the Dp427m promoter in the same tissue. Combined with these data and the observations that the Dp427l promoter resides in the intron of a sense-strand multi-exon gene and that Dp427l sequences are not conserved in mouse, Wheway & Roberts (2003) conclude that the Dp427l (lymphocyte) promoter is not a biologically significant part of the dystrophin gene.
This isoform is expressed predominantly in neurons of the cortex and the CA regions of the hippocampus. It was first detected by (Nudel et al.) and uses a unique promoter/exon 1 (Boyce et al.), located about 130 kb upstream of the muscle promoter. The cortical promoter does not contain typical transcription elements and has no TATA-box. The transcript splices directly into the common exon 2 of Dp427m and has a similar lenght, i.e. 14 kb. Dp427c contains a unique N-terminal MED amino acid sequence, exchanging the MLWWEEVEDCY-start of Dp427m. The remainder of Dp427c is identical to Dp427m.
Boyce et al. determined the human genomic sequence, identified the transcriptional start site and the long-range physical map around the promoter. Total dystrophin expression levels in brain are in the range of 1-2% of those in muscle. Using RT-PCR, Dp427c expression is detectable in heart muscle but hardly in adult skeletal muscle (Muntoni, Bies). The Dp427c isoform is also known as brain dystrophin.
Dp427c is up-regulated in skeletal muscle, but not cardiac muscle, of XLDC-patients with mutations in the Dp427m promoter / exon 1.
Dp427p was first identified in Purkinje cells by using in situ hybridization with antisense oligonucleotides to brain sections (Gorecki et al.). The isoform initiates from a unique promoter/exon 1 which is located in the first intron of the Dp427m gene. The Purkinje promoter drives nearly all cerebellar dystrophin expression. The transcript splices directly into the common exon 2 of Dp427m and has a similar lenght, i.e. 14 kb. Dp427p contains a unique N-terminal MSEVSSD amino acid sequence, exchanging the MLWWEEVEDCY-start of Dp427m. The remainder of Dp427p is identical to Dp427m.
Holder et al. detected two alterntively spliced Dp427p transcripts in cortical brain and adult skeletal muscle and, at much lower levels, in adult cardiac tissue. The two alternatively spliced transcripts, designated here Dp427p1 and Dp427p2, differed by the insertion of 82 nucleotides directly flanking Dp427p exon 1. The 82 nucleotides insertion in Dp427p2 introduces a translational stop codon 24 bp downstream of the ATG-codon used in Dp427p1; it is unclear whether any protein is generated from this transcript. Dp427p1 is the predominant transcript in fetal brain, Dp427p2 in adult brain. Fetal skeletal muscle contains low, but equal levels of Dp427p1 and Dp427p2 while adult skeletal muscle contains mainly Dp427p1 (Holder et al.).
Dp427p is up-regulated in skeletal muscle, but not cardiac muscle, of XLDC-patients with mutations in the Dp427m promoter / exon 1.
The Dp260 isoform was identified by Pillers et al. when they analysed the retina for dystrophin expression after observing abnormalities of retinal function by electroretinography, i.e. a reduced amplitude for the b-wave in the dark-adapted state. Dp260 expression in retina is found in the outer plexiform layer exclusively and lies at 10-20% of that in muscle.
Expression of Dp260 has also been found in brain and cardiac muscle. Transcripts originate from a promoter/exon 1 sequence located in intron 29 of the dystrophin gene which is alternatively spliced (White,R.A. GenBank accession number U27203). This alternative splicing creates two Dp260 isoforms, Dp260-1 and Dp260-2. Dp260-1 contains a 95 bp exon 1 encoding a unique 16 amino acid N-terminal MTEIILLIFFPAYFLN-sequence exchanging amino acids 1-1357 of Dp427m. Dp260-2 splices later, after 238 bp (in "intron 1"), giving a more downstream initiation codon and a 13 amino acid MSARKLRNLSYKK N-terminus. In mouse, a Dp260-2 isoform has been described with an identical N-terminus (D'Souza et al.).
Like other isoforms, Dp260 is alternatively spliced at its 3'-end, although exon 78 seems to be always present in retina (Pillers). Next to Dp260, retina also contains the Dp427m-, Dp427c- and Dp71-isoforms. Using RT-PCR analysis, Dp260 expression was also detected in murine brain and cardiac tissues but not in kidney, liver, lung, muscle, pancreas, spleen, testis and thymus (D'Souza). The Dp260 isoform is also known as R-dystrophin.
Dp140 was discovered by Lidov et al., analysing Western blots of micordissected brain regions. Dp140 expression was detected throughout the central nervous system (i.e. cerebral cortex, cerebellum, hippocampus, brain stem, spinal cord and olfactory bulb) and kidney but not in skeletal muscle, cardiac muscle, lung, liver and spleen (Lidov). Dp140 was detected in mdx-mice but not in mdx3Cv-mice, indicating that it is derived from the dystrophin gene and not a cross-reactive protein.
The Dp140 transcript originates at a promoter/exon 1 located in intron 44 of the dystrophin gene (Lidov). On a Northern blot, a Dp140 exon 1 probe detects a strong 7.5 kb and a weaker 10.5 kb band. Dp140 seems to have a long 1 kb 5'-UTR since translation is initiated in exon 51 (at amino acid 2461 of Dp427m). Unlike other isoforms, Dp140 thus has no unique N-terminus.
Lidov performed a detailed study of the Dp140 expression, splicing and splicing levels. Expression of the unique Dp140 first exon was only detectable in brain (cerebellum) and kidney. Differential splicing was detected for exons 71, 71-74, 78 and combinations of these producing at least five Dp140-isoforms.
Dp140 might be involved in mental retardation which is frequently found associated with DMD/BMD Emery et al..
The Dp116 was detected by (Byers et al.), using an antibody against the distal rod domain of dystrophin. Dp116 has been exclusively found in adult peripheral nerve, along the Schwann cell membrane (on Northern blots liver, brain, testes, lung, spleen, submaxillary gland, thyroid and kidney were negative). In these cells, no full-length nor Dp71 isoforms can be detected. Transcripts measure 5.2 kb and derive from a promoter/exon 1 which has been mapped in inron 55 of the Dp427m gene. Dp116 contains a unique N-terminal MLHRKTYHVK amino acid sequence, exchanging amino acids 1-2739 of Dp427m. The Dp116 isoform is also known as S-dystrophin and apo-dystrophin-2.
This isoform was first detected in liver by Bar et al., encoded by a 4.8 kb mRNA (originally described as 6.5 kb). The promoter/exon 1 is located ~8 kb upstream of exon 63 of the Dp427m gene (Rapaport et al.). Dp70 contains a unique N-terminal MREQLKG amino acid sequence, exchanging amino acids 1-3075 of Dp427m. It shares with Dp427m most of the sequence of the cysteine-rich and C-terminal domains. The originally described Dp71 cDNA did not contain dystrophin exons 71 and 78; Austin et al. showed that Dp71 can also be found with these exons. The absence of exon 78 causes a frame shift which replaces the 13 C-terminal dystrophin amino acids (RNTPGKPMREDTM) with 31 new ones (HNVGSLFHMADDLGRAMESLVSVMTDEEGAE). Isoforms containing this alternative C-terminus can be detected with specific dystrophin antibodies, like 462B.
Dp71 is ubiquitously expressed, but undetectable in fully differentiated skeletal muscle. Dp71 is the most abundant dystrophin in brain and liver and the predominant isoform in astrocyte and glioma cell cultures. The amount of mRNA is in some tissues comparable to the amount of dystrophin mRNA found in muscle (Rapaport et al.). Dp71 mRNA is alternatively spliced: varying degrees of mRNA +/- exon 71 and +/- exon 78 are found in specific tissues generating four Dp71 isoforms (Austin et al.). The Dp71 isoform is also known as apo-dystrophin-1.
Dp40 is a dystrophin isoform produced by the use of an alternative poly-A additon site.
Alternative splicing of dystrophin RNA is extensive and it creates a range of dystrophin isoforms with only small differences. It is often very difficult to discriminate between these isoforms and only when is a specific tissue or cell line only one isoform is expressed one can be absolutely sure that a specific type of alternative splicing is occuring in that transcript. Alternative splicing was first reported by Feener et al., studying human brain and muscle samples. The spliced forms reported included -exon 68, -71, -78, -71-78, -71-72-78 and -71-72-73-74-78.
The NOMENCALTURE we use for the alternatively spliced transcripts is: a-types miss the exon 71 sequences, b-types the exon 78 sequences and c-types the exon 71-74 sequences. The b-types have an alternative 31 amino acid C-terminus against which specific dystrophin antibodies (like 462B) could be generated.
Surono et al., studying the region between Dp427m exon 1 and exon 18, identified six alternatively spliced transcripts in the 5' region of the gene.
Lidov et al. performed a detailed study of the Dp140 expression and splicing. They detected five Dp140-isoforms; Dp140 (1 in 9 clones in kidney), Dp140c (missing exons 71-74, 4/12 clones in cerebellum and 1/9 in kidney), Dp140b (missing exon 78, 6/9 clones in kidney), Dp140ab (missing exons 71 and 78, 5/12 clones in cerebellum) and Dp140bc (missing exons 71-74 and 78, 3/12 clones in cerebellum and 1/9 in kidney).
Austin et al. published an extensive study on the differential splicing of exons 71 and 78 in Dp71. They cloned all possible transcripts, with and/or without exons 71 and 78, from human amniotic fluid cells. Detailed analysis showed that brain, kidney, liver, lung, muscle and testis contained Dp71-transcripts with and without exon 71 while heart had only 71+ transcripts. Dp71 exon 78 + or - transcripts were found in brain, kidney, lung, muscle and testis, while only 78+ transcripts were found in heart and liver. The alternatively splicedtranscripts encode four different protein isoforms; Dp71, Dp71a, Dp71ab and Dp71b.
Analysis of ectopic (or 'illegetimate') dystrophin transcripts, mostly derived from peripheral blood lymphocytes (PBL), is frequently used in RNA-based mutation detection. For such samples, alternative splicing, i.e. skipping of the exon, has been reported for exons 9 (Reiss & Rininsland), 38, 68, and 74 (Roberts/Gardner). In addition, Tuffery et al. reported skipping in PBL RNA of exons 14+15, 39, and 48 and Barbieri et al. of exon 25.
Although most forms of alternative splicing reported thusfar concern the skipping of exons, Roberts et al. have reported the inclusion of an exon 'X' sequence between Dp427m exons 1 and 2 blood-derived RNA. The exon 'X' insert could not be identified in muscle RNA.
The normal 3'-terminal exon present in mRNA's derived from the dystrophin gene is exon 79. The use of an alternative polyA-addition site, localized in intron 70 of the dystrophin gene, was first reported by Feener, studying human brain and muscle samples. The new 3'-terminal sequence would generate a translational stop codon in the splice donor site of exon 70. The encoded protein would miss 277 amino acids compared with the full-length Dp427m (about 32 kD) and could represent a ~405 kD protein reported to be present in mouse smooth muscle.
Dp40 was first identified by Tinsley et al. after sequencing of a cDNA clone isolated from a rat Schwannoma library. Dp40 is encoded by an 2.2 kb mRNA. The 5'-UTR and first 7 amino acids are identical to that of the 4.8 kb mRNA encoding Dp71, but the 3'-UTR derives from intron 70 sequences. The stop codon of this isoform lies directly at the splice junction of the exon/intron 70 boundary of the dystrophin gene. The use of an alternative poly-A addition site localized at this position was previously detected by Feener et al. in a truncated fetal dystrophin. Dp40 has an estimated length of 341 amino acids and a molecular weight of 40 kDa. Dp40 is unique since it lacks the normal C-terminal end of Dp427m (amino acids 3409-3685). The tissue distribution of the Dp40 transcript is similar to that of Dp71, although it is less abundant. Using RT-PCR, Tinsley et al. detected expression in all human and mouse tissues examined (i.e. fetal muscle, fetal lung, fetal liver, fetal brain, adult muscle and a rat Schwannoma cell line). In murine ES-cells, Dp40 was the only detectable dystrophin expressed. Dp40, which was not visualized on Western blots, is also known as apo-dystrophin-3.
Since many different dystrophin isoforms exist, it is very difficult to identify one specific form. In immunohistochemistry, most antibodies detect several isoforms and in RNA PCR many primer pairs amplify several isoforms. Furthermore, every time a new isoform is identified, one has to check if previous experiments were conclusive in discriminating between an isoform studied and the newly detected isoform.
Specific detection of dystrophin isoforms on protein level is simple on Western blots, where the size of the detected protein is usually conclusive (see also DMD antibodies).
For specific detection of dystrophin isoforms on RNA-level two methods have been described:
Several reports demonstrate unexplained consequences of mutations on the expression of the different dystrophin isoforms and their balance. The most prominent example is found in some cases of X-linked dilated cardiomyopathy (XLDC). Several Dp427m promoter / exon 1 mutations have been reported in which expression in muscle is taken over by the Dp427c and/or Dp427p promoter but not in heart (see XLDC homepage).
| Top of page | LMDp home page
|
| dystrophin associated proteins | dystrophin sequenes |
| Remarks / information | Copyright©,
liability |