McNaughton, J.C., Cockburn, D.J., Hughes, G., Jones, W.A., Laing, N.G., Ray, P.N., Stockwell, P.A., and Petersen, G.B. (1998). Is gene deletion in eukaryotes sequence-dependent? A study of nine deletion junctions and nineteen other deletion breakpoints in intron 7 of the human dystrophin gene. Gene 222, 41-51

Positions are those of the 110kb DNA sequence of intron 7 - Genbank Accession No U60822 - McNaughton et al. (1997) Genomics 40, 294-304.
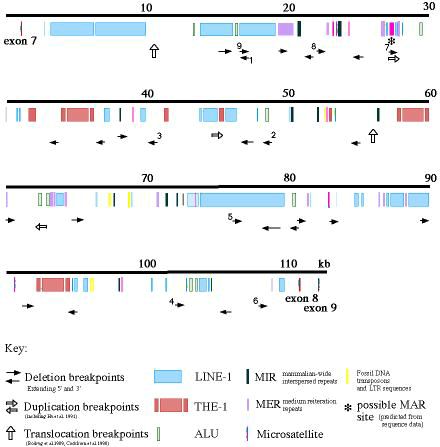
| Name | Length | F Position | Forward Primer | R Position | Reverse Primer |
|---|---|---|---|---|---|
| exon 7 | 127 | 1060 | AGCAGTCAGCCACACAACGACTGG | 1186 | CTCTACCATACTAAAAGCAGTGG |
| 11k | 269 | 11429 | TCCCTGCATCACTCTTGAG | 11697 | ATGTGATGATTACAGCAGAG |
| 14k | 297 | 13803 | GGAAACCTGAAGATTTATAG | 14100 | AAGCAGAAAGGGGTGACTAGAG |
| 14.4k | 1219 | 13803 | GGAAACCTGAAGATTTATAG | 15021 | GGGACTGTAGAAAATGAAAG |
| 14.8k | 2120 | 13803 | GGAAACCTGAAGATTTATAG | 15922 | TAGAAGAAATCGTTGTCC |
| 15k | 2578 | 13803 | GGAAACCTGAAGATTTATAG | 16380 | GTATCAAACTGAATGGGG |
| 16k | 636 | 16321 | CCTGACCATTCCAGTTTTATAG | 16956 | GACACATACAACCAACCGTGG |
| 18k | 1635 | 17363 | GTAGGTGTTATTATATTTTG | 18997 | GAATCTCAGAACATGAAAACAGG |
| 19k | 151 | 19247 | GAGCTGTGTACATTGGCATTG | 19397 | ATTACCCGCACCATGCTTGCTG |
| 19.2k | 1178 | 18597 | TCACTGTTAATCTGATGGAGG | 19774 | CTGCCTTCCACCTGAGCAT |
| 19.6 | 345 | 19430 | GCAAGCAATCCATGATGG | 19774 | CTGCCTTCCACCTGAGCAT |
| 19.7 | 701 | 19430 | GCAAGCAATCCATGATGG | 20130 | AACATGGCCTGGTTTTGAGAG |
| 20 | 1114 | 19430 | GCAAGCAATCCATGATGG | 20543 | TCTGGGGCTTCAAAATGGCTG |
| 20.1k | 315 | 20568 | GGAGTTAGTGGATCATATGG | 20882 | ATTATCTAGAACATGACTGCG |
| 22k | 1574 | 21273 | GTCCCAGCATTTGTTTAG | 22846 | GGACTCCTGTATTTAATTTG |
| 22.1k | 892 | 21955 | GGGTTAGTTTTGCTAAACAG | 22846 | GGACTCCTGTATTTAATTTG |
| 23k | 175 | 22672 | GTATTTGGCCCGTAATATGAG | 22846 | GGACTCCTGTATTTAATTTG |
| 24k | 232 | 23991 | TGGATGCACATCCATCAGTTG | 24222 | GGAATATTTTCATAACTGGTG |
| 25k | 193 | 24566 | GGCAAGAGTACATGGTGAAG | 24758 | AAAACCTAGTTACTTCACCAG |
| 26k | 1237 | 25456 | TGTGCAGGTGGCTGAGTAG | 26692 | ACAGAGATTTCCCGATCCAAG |
| 26.6k | 167 | 26526 | TTCTGCTCTTTAATGGCTGAG | 26692 | ACAGAGATTTCCCGATCCAAG |
| 27k | 497 | 27213 | GTGGGTACATACTAGGGGTG | 27709 | CCATGTACCCACAAAAATTA |
| 27.5k | 747 | 27213 | GTGGGTACATACTAGGGGTG | 27959 | TGCAAATTCAAATAACCTCTG |
| 28k | 175 | 28203 | GATGTTGTTCTTCCACTCTG | 28377 | GACTTGGTGTGTATAGTATG |
| 29k | 113 | 29379 | TCACAATGCTGTAGGTAGTTG | 29491 | AGGCACTCTTATGCTCTGG |
| 30k | 422 | 29379 | TCACAATGCTGTAGGTAGTTG | 29800 | CAGGCACTCGCACTTTCAGAG |
| 32k | 457 | 32214 | AGCTCTGGTCTGTTGTACTTGGCTG | 32670 | CCATACAAGAAGCAATCCCAGCAAG |
| 34k | 544 | 33800 | AAGCATTTGAGAACATCAGTGG | 34343 | TGCCAGTTACCCAGTTCCAAAG |
| 34.1k | 458 | 33886 | TTATTCTGCATTAATAAGTTG | 34343 | TGCCAGTTACCCAGTTCCAAAG |
| 35k | 2461 | 33800 | AAGCATTTGAGAACATCAGTGG | 36260 | CTAGGTAGTTCTATCACTTAG |
| 36k | 447 | 35814 | GAAGGCATGACTGTGTTTTG | 36260 | CTAGGTAGTTCTATCACTTAG |
| 36.2k | 697 | 35814 | GAAGGCATGACTGTGTTTTG | 36510 | TAAAGGGCAAGGGTTCCAC |
| 36.8k | 1273 | 36240 | CTAAGTGATAGAACTACCTAG | 37512 | GACTGGAGGACATTACAGGTG |
| 37k | 710 | 36803 | TGTAGGAATAGATCAGATGCCAG | 37512 | GACTGGAGGACATTACAGGTG |
| 37.6k | 1236 | 36803 | TGTAGGAATAGATCAGATGCCAG | 38038 | TACTCTTACATAGAGGTAG |
| 37.7k | 1294 | 36803 | TGTAGGAATAGATCAGATGCCAG | 38096 | TCTGGTAGGTAGCAGAGCTAG |
| 39k | 157 | 38785 | GTTGAGCCCATCCTTGAG | 38941 | CCACTTTTGCTATTTCAGAGG |
| 39.2k | 176 | 39166 | GGGAATTTGTCAATGATGAG | 39341 | GATCATGCCAGCCTTAGAAAG |
| 40k | 1560 | 40005 | TGTATTCCCTCTCCAAATG | 41564 | GAAGGTAGAGCCACAACAAAGG |
| 41k | 98 | 40910 | CCACAGAATTTGAGGTGACAC | 41007 | ATAGAATTCAGTTGCACCAGGCCATCAAG |
| 41.3k | 594 | 40971 | TGATTAGTCTTGATGGCCTGG | 41564 | GAAGGTAGAGCCACAACAAAGG |
| 43k | 169 | 42732 | CATGGTGTATTGCACCTTTCTCCCAAACAG | 42900 | GATTCTACTGTCACAAGAACCGATG |
| 45k | 592 | 44989 | GCAACTTACGGAATGGGAGA | 45580 | CTACTCATGGTCTTACTGGCCA |
| 45.3k | 529 | 45052 | ACTGTGCCAAGAACTCCTATA | 45580 | CTACTCATGGTCTTACTGGCCA |
| 45.5k | 173 | 45475 | ATTATATCATCGTAGAATTCA | 45647 | TGGGTGTGAAGTGCTATCT |
| 46k | 800 | 45475 | ATTATATCATCGTAGAATTCA | 46274 | ACCTTGTTGTGTCACCATCC |
| 46.6k | 657 | 46334 | GTTATATATATTTTACCATG | 46990 | GTAATGGTCTACCAACCTTAT |
| 47k | 352 | 46639 | GTTATCAGTTTAGGAAGAATG | 46990 | GTAATGGTCTACCAACCTTAT |
| 48k | 215 | 47546 | GTTCATTGTGTACAGCGGTAG | 47760 | GAGAGACTGCTTAGGAAG |
| 50k | 168 | 49624 | AATGCCCGGAGAGTACATTTG | 49791 | TGTACCTCACATAATGACATG |
| 53k | 899 | 52493 | TCAGGAACATGTGGGACCTGG | 53391 | CACCCAACAGTTCTCTTGAATT |
| 54k | 223 | 53824 | GTAGATGGGGCAAGTGACAGC | 54046 | CCTTGTGGATCAATTTGGGAAAC |
| 55k | 837 | 54452 | GTTGAGCCGGAATGGTGAG | 55288 | TAAAGGAAGATACAGAAGTGAG |
| 55.2k | 185 | 55104 | TCCTGTGTTGGGCTGCTCTTAGTTGG | 55288 | TAAAGGAAGATACAGAAGTGAG |
| 58k | 556 | 57486 | GGAAGTGATATTTAGATGGAAGTCTGGG | 58041 | GACTTTGGTGCACTGTTGGGAAGGCAAG |
| 58.4k | 1711 | 57486 | GGAAGTGATATTTAGATGGAAGTCTGGG | 59196 | GGGGTGATTTAGCGTTATCTGGTG |
| 59k | 341 | 59568 | GAGAATGGATAATTTATACAGG | 59900 | CACCTATTTTACTTATACCTACTG |
| 59.9k | 179 | 59889 | GTAAAATAGGTGAAGGGTGG | 60067 | GCAGAGCCAAACTCCTAGGCAGATAGG |
| 60k | 494 | 59889 | GTAAAATAGGTGAAGGGTGG | 60382 | TATCCATCCTTGAGAAAGCTAG |
| 61k | 940 | 60890 | GGTGTCTACCTCAATTATCAG | 61829 | TGTCATCAGTCATTGCAGAG |
| 61.7k | 221 | 61609 | GTATGTACCTAGGACCTAATG | 61829 | TGTCATCAGTCATTGCAGAG |
| 62k | 290 | 62577 | GGCCTACACTTTTTCTTTAAGG | 62866 | GGGAGGAATCCGTTTTAA |
| 64k | 105 | 64131 | TGGGAGAGTCCTGAAATTCGG | 64235 | GAAGCAGTGGTCTAGGGCAG |
| 64.6k | 1038 | 64131 | TGGGAGAGTCCTGAAATTCGG | 65168 | GTAGGAATGATGGTTTCACAG |
| 66k | 264 | 66006 | ACAAGTGAACAGAAGTCCAG | 66269 | GGTCTTCCTAGCATTGTGTG |
| 72k | 408 | 71771 | GAGTGAATTTCACACCTCTCCTTTTG | 72178 | GAAGTTCCTAGCCCTTAATAAAG |
| 73k | 244 | 73351 | GGAGTTTGAAATCTGGATG | 73594 | GAGTAAGTTTTAATTAACAGG |
| 73.5k | 398 | 73351 | GGAGTTTGAAATCTGGATG | 73748 | ATGTACCCTAAAGCTTAAAG |
| 79k | 1832 | 77971 | GAAGCTTAGTTTGCCTGGATG | 79802 | GGAATAGAGCCTACCAGTGG |
| 79.7k | 84 | 79679 | GTCGCTCACTCTGGGAGCTG | 79762 | TTGGATCAGAAATCTGATTTG |
| 80k | 440 | 79679 | GTCGCTCACTCTGGGAGCTG | 80118 | GTCTTCCTAGGCAATAGGAATG |
| 80.4k | 352 | 80260 | TACTTATGGCACCATTACATT | 80611 | CCCCACTGTTAAAATGTAACGT |
| 80.5 | 1048 | 80235 | GGATTGGATCACATATCTATGC | 81282 | TGCCCAATATTTAGTCTTGAG |
| 80.6k | 1023 | 80260 | TACTTATGGCACCATTACATT | 81282 | TGCCCAATATTTAGTCTTGAG |
| 80.9k | 691 | 80592 | GTTACATTTTAACAGTGG | 81282 | TGCCCAATATTTAGTCTTGAG |
| 81k | 149 | 81134 | TGCCAAGCTACCATGAATCAG | 81282 | TGCCCAATATTTAGTCTTGAG |
| 82k | 238 | 82023 | GTCTGATAAGACAAGCTTTGTTG | 82260 | GTGAGGTCTATATTCAAATG |
| 83.4k | 410 | 83195 | GGTGGTCTAGTCAGTTGCTTG | 83604 | GGTTGAAGTAAAATCAATGTG |
| 83.5k | 679 | 83195 | GGTGGTCTAGTCAGTTGCTTG | 83873 | CAGACATTGGATGTGCTAATT |
| 83.6 | 801 | 83195 | GGTGGTCTAGTCAGTTGCTTG | 83995 | GGAAAGAGCAGTTATAAG |
| 84k | 132 | 84501 | TAGAAAGTTAAGATTGCGTAG | 84632 | GGAATATATGCAATTTCTAATAGG |
| 86k | 901 | 85613 | GGCTAAGGTGTTGTACTTGAGG | 86513 | GGTGCAATTCTACTTCTTGAGG |
| 87k | 532 | 87588 | GGTTGTGGGGTGGTTGTACTG | 88119 | GATACCAAGGGACAGCTAT |
| 88k | 804 | 87588 | GGTTGTGGGGTGGTTGTACTG | 88391 | GGATGCAACTGTAGACCAGAA |
| 89k | 1127 | 88280 | TGCTATAGAAATAGTTGTTACCAT | 89406 | GGGATTGTTGGGTCATATGGTAG |
| 89.8k | 178 | 89698 | AATGTATAAGTTGAATACATAG | 89875 | TTTACCCCATTATTATCTG |
| 90k | 213 | 90069 | ATTCAACACCCATGGACCAAG | 90281 | ACATCTCATTGCTCTTCTTAG |
| 91k | 254 | 90978 | TCAAAGCATCATGTGCATCTG | 91231 | GTGTAGGGTGATGAAGAATG |
| 92k | 1117 | 90978 | TCAAAGCATCATGTGCATCTG | 92094 | ATACACATATCAAGATAAG |
| 94.3k | 492 | 94033 | TCATGTATCTTCGTAGCAGTG | 94524 | GAACAGCGTCAATACAGAG |
| 94.5k | 76 | 94449 | AAATCATATCAATATGCTAG | 94524 | GAACAGCGTCAATACAGAG |
| 95k | 1737 | 94686 | TCAGCATAGTTACTTTTTG | 96422 | TCCTCTCTAGATTATTTCTTG |
| 95.5k | 1283 | 95140 | GTGTATCTTATAGCATCATG | 96422 | TCCTCTCTAGATTATTTCTTG |
| 96k | 169 | 96254 | GTATGTATGCGACTCTCTGTG | 96422 | TCCTCTCTAGATTATTTCTTG |
| 100k | 838 | 99667 | AGGAAGCTGAGGTGCATTATG | 100504 | GACCCAATCCTTCACACTTAG |
| 100.3k | 1373 | 99667 | AGGAAGCTGAGGTGCATTATG | 101039 | GGACGAACATCACGCATTTGTG |
| 101k | 2006 | 99667 | AGGAAGCTGAGGTGCATTATG | 101672 | GAGCTTAGCCTGTTGAGG |
| 102k | 240 | 102656 | GAAGAGGGCTTTTATGGAAG | 102895 | ATTGAAAATATGAGACACAG |
| 103k | 131 | 103249 | GGATTGGCTGCATGATAAG | 103379 | GAGCATAACAGGGAAAAAAG |
| 104k | 190 | 104480 | GAGCCACTGTTTTCTACTTG | 104669 | TGCGTTGATGAAGATGATTG |
| 105k | 1241 | 105222 | GGCTAGTTTATGACCAGAAG | 106462 | TATGAGTATATAGCAGATGAG |
| 106k | 184 | 106279 | GACTTTAACTGTCCTCTG | 106462 | TATGAGTATATAGCAGATGAG |
| 107k | 1088 | 106279 | GACTTTAACTGTCCTCTG | 107366 | GGATTGCTGGAGGAAAACTTG |
| 108k | 520 | 107761 | ACTGGTTGTGCCAATTAGAGG | 108280 | GACACATGCATGTATGTTTGG |
| 109k | 213 | 108664 | GTGTAATAAGTTGAGCCATATG | 108876 | GGAGCTCTGCATTTGCAATAG |
| exon 8 | 361 | 110557 | AGGCCTCATTCTCATGTTCTAATTAG | 110917 | GTCCTTTACACACTTTACCTGTTGAG |
| Name | Location | Forward Primer | Location | Reverse Primer |
|---|---|---|---|---|
| Junction 1 | exon5F | GGTTGATTTAGTGAATATTGGAAGTACTG | intron7R @19003 | ACAGAAGAATCTCAGAACATGAAAACAGG |
| Junction 2 | intron5F | GGAATGAGGCTTTCTCAGTCTTGGG | intron7R @45512 | TGCTGTTTTTCTGTTGTTGAATTCTACGATG |
| Junction 3 | intron5F | GGAATGAGGCTTTCTCAGTCTTGGG | intron7R @41095 | GGCACTTATGTGTTAAAGCAGCAAGG |
| Junction 4 | intron7F @100853 | GGTCCTAATTACAATGGTGGAAGCCAG | exon10R | GGTCCAGGTTTACTTCACTCTCCATCAATG |
| Junction 5 | intron7F @73424 | GTCTTGAAGCACCAGGTGGCCATTTGGGCTGAG | exon12R | CTTATGTTGTTGTACTTGGCGTTTTAGG |
| Junction 6 | intron7F @107316 | GGGATTATGGTCCTAGCCTCCTATTTCT | exon16R | CACCGGGCAAAGTTATCCAGCCATG |
| Junction 7 | intron7F@27213 | GTGGGTACATACTAGGGGTGTGTGTG | exon17R | GGGGAGGTGGTGGTGGAAGTTCCTCTTG |
| Junction 8 | intron7F @20494 | GGAAGAGGTGAGTAACAGATCCATCAATTCAG | exon21R | GTCTGCATCCAGGAACATGGGTCCTTG |
| Junction 9 | intron7F @16318 | ACGCCTGACCATTCCAGTTTTATAGGGAG | exon25R | ACCTGTTGGCACATGTGATCCCACTGAG |
| Dup Junction* | intron7F @62577 | GGCCTACACTTTTTCTTTAAGG | intron7R @45629 | TGGGTGTGAAGTGCTATCT |
*Duplication junction in intron 7 associated with deletion junction 2